Help
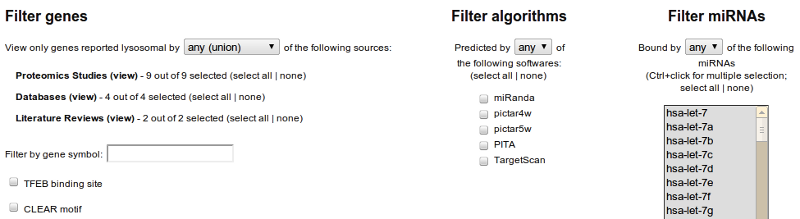
Two types of queries can be made:
- Genes: To find lysosomal genes, ignore the two columns on the right (algorithms and miRNAs). You may specify the sources (studies and databases) which should be considered, either in union of intersection (by default, the union of all sources is selected). In addition, you may filter by gene symbol (or part thereof), or display only genes which have a TFEB binding site or a CLEAR motif near their transcription start site.
The result of the query is a list of genes matching your criteria. You can get more information on each of them (and the miRNAs predicted to bind on their transcript) by clicking on the gene's symbol. - Bindings: To see which miRNAs are predicted to bind on which genes' transcripts, you must select at least some of the miRNAs (rightmost column) and some of the prediction softwares (middle column). You may also filter the genes as per gene queries.
The result of the query is a list in which each line represent a putative binding between a specific miRNA and any transcript of a specific gene. By clicking on any gene, you can get additional information about the positions of the bindings and their transcript specificity.
The interface to the database requires javascript to be activated.
Sources & Softwares used for the predictions
miRanda
Target Site Predictions, Good mirSVR score, Conserved miRNA, August 2010 Release Downloads
TargetScan
Predicted Conserved Targets Info
PITA
PITA Sites catalog, Top, no flank
picTar 4-5 way
Predictions as published in Lall et al. 2006.
TFEB binding sites
sites as reported in Palmieri et al. 2011.
CLEAR motifs
Positions on human genome (hg19) of the CLEAR sequence "GTCACGTGAC" searched using fuzznuc allowing a single mismatch.